Benchmarking Biointelix Modeling Against Published Experimental Structures
The purpose of this internal study was to evaluate the accuracy and reliability of Biointelix’s bioinformatic modeling and docking workflow by comparing its predictions with previously published experimental data. As a test system, we selected the interaction between Green Fluorescent Protein (GFP) and a specific nanobody, Lag16, which has been structurally characterized by Zhang et al., 2020 using X-ray crystallography.
In this benchmark, we reproduced the entire interaction modeling process starting from the amino acid sequences of GFP and Lag16. Our workflow included structure prediction, molecular docking, and interface residue analysis, integrating the Biointelix-developed scoring formula for interaction ranking. The published X-ray structure and interaction data from Zhang et al. served as positive controls to assess the performance and biological relevance of our computational predictions.
The comparative analysis demonstrated that our bioinformatic approach successfully identified several key interface residues that were also reported by Zhang et al. based on experimental data. These overlapping residues confirm the consistency and predictive accuracy of the Biointelix modeling platform in reproducing experimentally validated protein–protein interactions.
A summary table will present the residues in common between the experimental and Biointelix-predicted models, illustrating the alignment between computational docking results and crystallographic findings.
Experimental and Computational Analysis of Nanobody–GFP Interaction (residues identified in at least two models, see table)

Computational Design of Multi-Epitope Vaccines Against Influenza A
The objective of this project was to design next-generation, broadly protective vaccine candidates against Influenza A (H5N1) using a fully in silico bioinformatic and structural modeling pipeline. We aimed to demonstrate how computational tools can accelerate vaccine discovery and guide experimental prioritization.
We began by analyzing more than 20,000 H5N1 and 190,000 Influenza A HA sequences to identify conserved and immunogenic regions. Based on this analysis, we developed three vaccine constructs:
EpitoCore-HA-VX — a multi-epitope fusion including CTL, HTL, and B-cell epitopes from conserved HA regions.
StructiRBD-HA-VX — a rationally designed construct preserving the receptor-binding domain structure and incorporating an adjuvant sequence with optimized linkers.
FusiCon-HA-VX — focused on the highly conserved fusion peptide region to promote cross-subtype immunity.
Two comparator HA structures from published sources were included under the same modeling workflow. Structural models were generated from amino acid sequences, refined, and analyzed for antigenicity, allergenicity, toxicity, and population coverage.
All three Biointelix vaccine constructs, as well as the comparators, elicited robust immune responses in silico when tested using immune simulation tools, suggesting their potential to induce both humoral and cellular immunity. Structural alignment showed that the designed vaccines retained native-like conformations while effectively exposing epitope regions for immune recognition.
Conclusion
This study demonstrates how computational vaccine design can provide valuable preliminary insights into immunogenic potential, safety, and structural properties before moving into laboratory validation. While in silico results do not replace experimental studies, they serve as a powerful screening and optimization tool that can significantly accelerate early-stage vaccine research and reduce experimental costs.
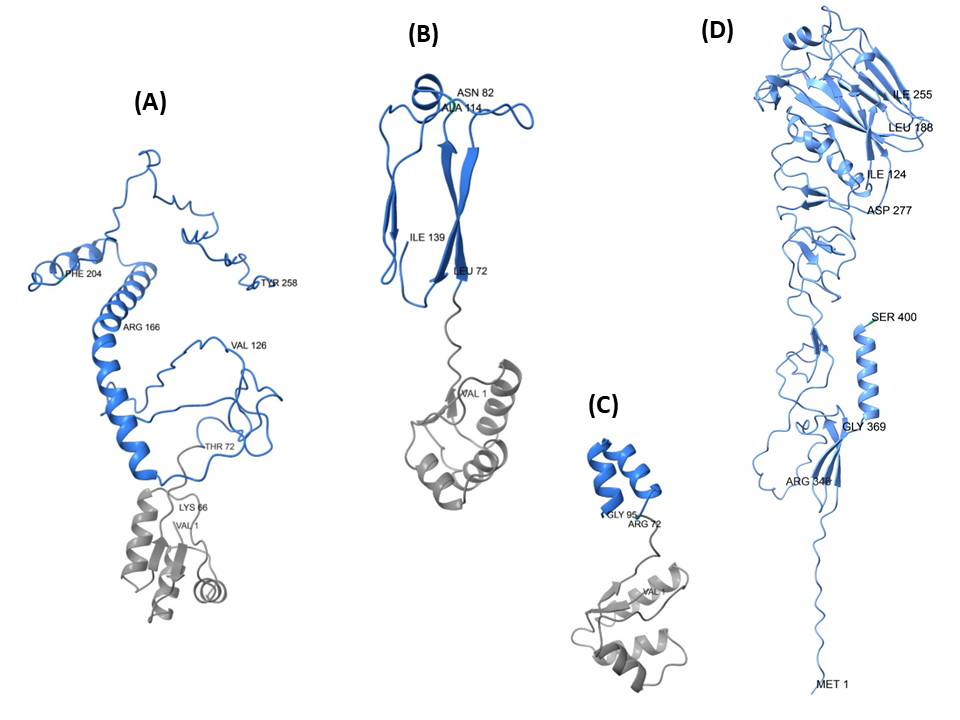
Predicted 3D Models of the Designed Influenza A HA-RBD Vaccine Candidates.
Ribbon diagrams display the AlphaFold-predicted tertiary structures of the three Biointelix vaccine constructs: (A) EpitoCore-HA-VX, (B) StructiRBD-HA-VX, and (C) FusiCon-HA-VX. Panel (D) depicts the reference HA fragment (residues 1–400 of H5N1 HA) used as a structural comparator. In each model, the blue segments correspond to receptor-binding domain epitopes, while the gray regions represent the incorporated adjuvant elements. The N- and C-terminal positions are indicated, along with selected residues highlighting key functional or structural features of the constructs (Palma et al., 2025).
