Antibody structure prediction
Introduction to Antibody Structure
Antibody structure prediction plays a critical role in therapeutic antibody engineering, vaccine design, and diagnostic development. Specifically, B cells produce antibodies (immunoglobulins), Y-shaped proteins that recognize and neutralize pathogens. Importantly, the variable domains (VH and VL), especially the complementarity-determining regions (CDRs), form the antigen-binding site and define binding specificity.
Nanobodies (single-domain antibodies or VHH domains) represent a unique subclass derived from camelid heavy-chain-only antibodies. In contrast to conventional IgG antibodies, nanobodies contain a single variable domain, which makes them smaller, more stable, and easier to engineer. As a result, researchers now use nanobodies extensively in drug discovery, imaging, and targeted therapies.
Accurate antibody structure prediction forms the foundation of modern therapeutic design, vaccine development, and immunodiagnostics. At Biointelix, we therefore generate high-quality 3D antibody structures using advanced computational modeling. In addition, researchers can explore experimentally determined antibody structures through the Protein Data Bank (PDB) and obtain sequence and functional information from UniProt. For example, IMGT provides a comprehensive resource for curated antibody data, while the Structural Antibody Database (SAbDab) offers validated 3D structures to refine predictions. Furthemore, researchers can perform antibody docking studies using SwissDock to evaluate antibody-antigen interactions in silico.
Methods for Antibody Structure Prediction
We apply tailored computational approaches to predict antibody and nanobody structures. Specifically, these include:
- Homology Modeling – Leverage structural templates from the Protein Data Bank (PDB). Additionally, AI-based tools enhance prediction accuracy by learning from large datasets.
- AI-Based Prediction – Use AlphaFold and IgFold, trained for immunoglobulin folds.
- CDR Loop Modeling and Refinement – We improve the accuracy of antigen-binding loops.
- Docking and Complex Prediction – Model antibody-antigen interfaces to assess binding.
- Stability and Developability Analysis – We evaluate solubility, folding, and expression in silico.
Antibody Structure Prediction Services at Biointelix
We deliver customized workflows based on antibody type and application. For instance:
- scFv Structure Prediction – Generate high-quality models for single-chain variable fragments.
- Fab and IgG Structure Prediction – Model frameworks with accurate VH/VL pairing.
- Nanobody (VHH) Structure Prediction – Specialized workflows for single-domain antibodies.
- Furthermore, our CDR Loop Refinement improves the accuracy of antigen-binding loops.
- In addition, antibody-antigen docking models binding sites and epitope recognition.
- Mutation and Stability Analysis – Predict effects of substitutions on binding and folding.
Moreover, Biointelix predicts the 3D structure of nanobodies (VHHs), single-chain variable fragments (scFvs), and fragment antigen-binding (Fab) domains directly from amino acid sequences with AlphaFold. We then refine models using GalaxyRefine to ensure accuracy and stability.
Example Structures
Three-Dimensional Structure of a Nanobody (VHH Domain)

Structural Representation of an scFv fragment
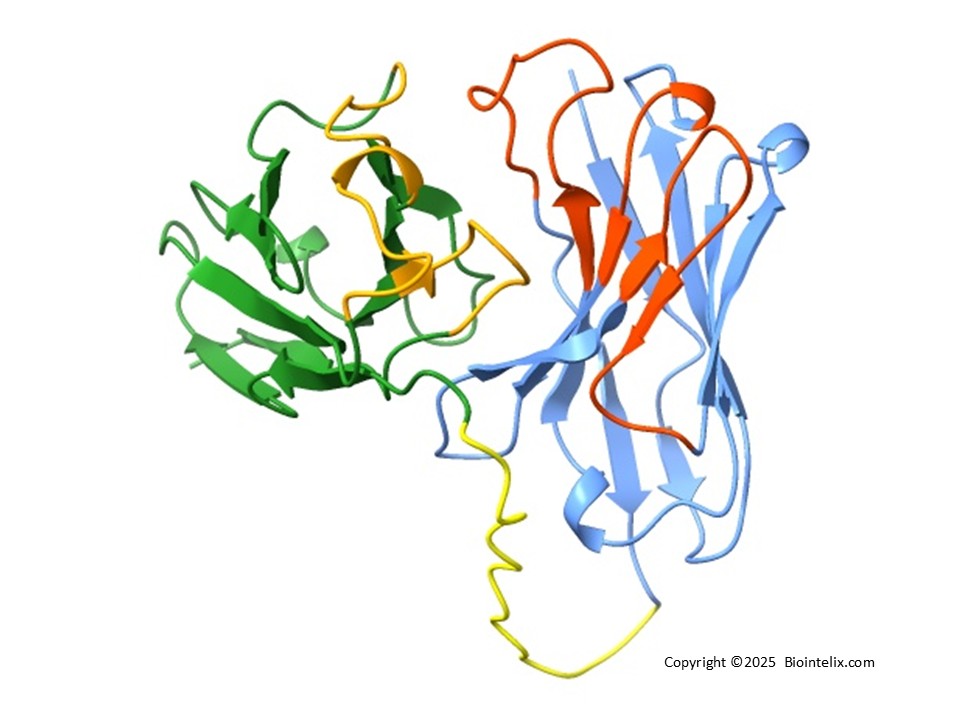
Molecular Structure of a Fab Fragment
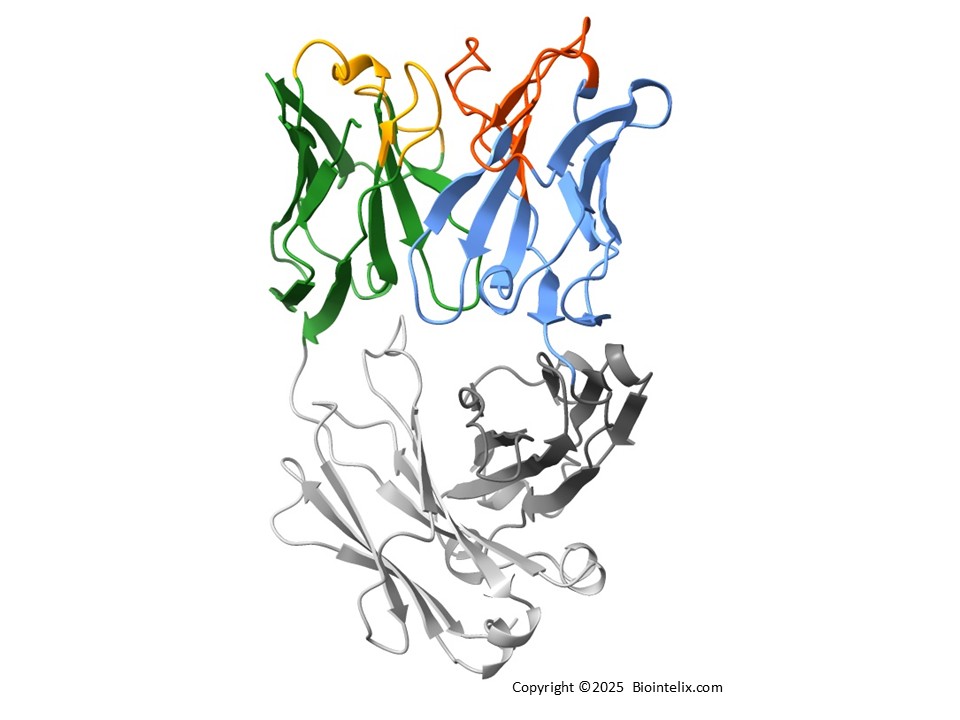
Why Choose Biointelix
- Firstly, we deliver publication-ready models for research, patents, and clinical development.
- Secondly, we provide end-to-end in-silico protein modeling with refinement for high-quality results.
- Finally, we offer fast, secure, and customizable solutions tailored to your research needs.
Get Started
Biointelix provides accurate, fast, and reliable antibody structure prediction services, from conventional IgGs to nanobodies. Consequently, whether your goal is drug discovery, vaccine design, or diagnostic development, our models deliver actionable insights.
Contact us today to discuss your antibody modeling project.
