Protein Structure Prediction Services by Biointelix
At Biointelix, we provide advanced Protein Structure Prediction Services that empower researchers, biotech startups, and pharmaceutical companies. Through AI-based protein structure prediction services and 3D protein structure prediction services, we accelerate drug discovery, vaccine design, and protein engineering. Consequently, our solutions deliver accurate computational insights without the cost and delays of traditional lab experiments.
Example: scFv Protein Structure Prediction
For instance, we recently modeled a Single-Chain Variable Fragment (scFv) antibody from its amino acid sequence using AlphaFold, followed by expert refinement and validation. As a result, the highest-quality model was selected based on structural integrity and confidence scores.
Structure Prediction of a Single-Chain Variable Fragment against spike protein
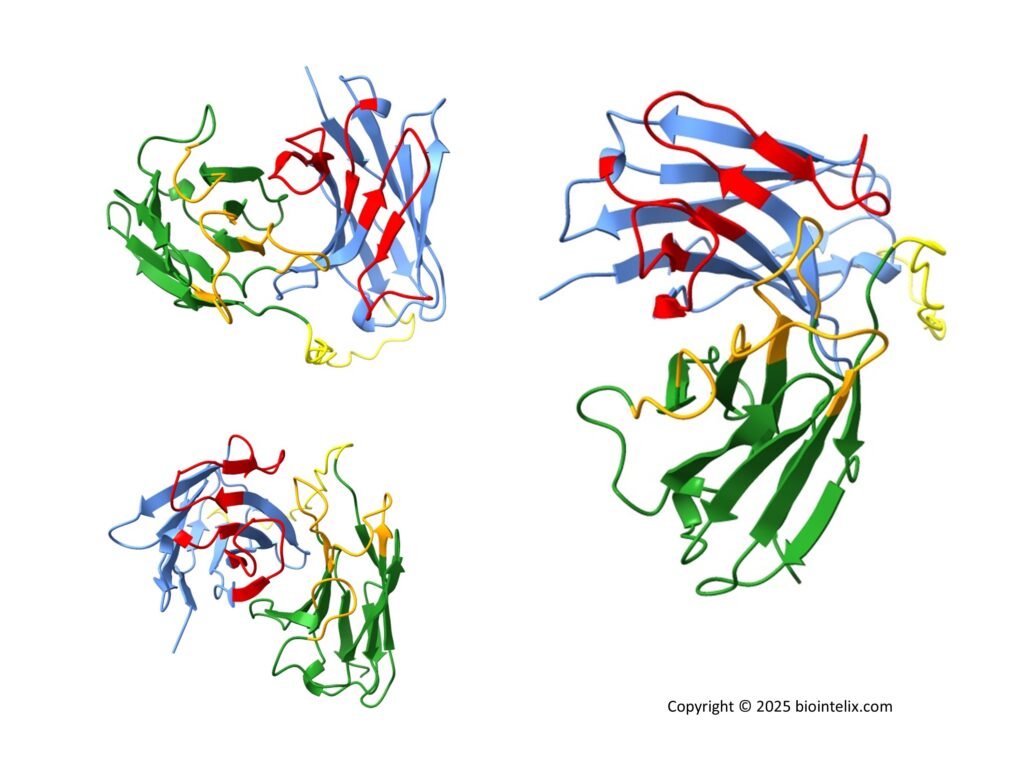
Interactive 3D Structure of scFv
In the 3D model:
- VH domain: blue
- VL domain: green
- Linker region: yellow
This example clearly shows how AI-based protein structure prediction services, combined with expert refinement, deliver reliable and actionable structural insights for antibodies and other protein targets. Moreover, we validate our protein models using structural databases such as the Protein Data Bank (PDB).
Model Validation & Quality Assessment
At Biointelix, accuracy is our priority. Each predicted structure undergoes rigorous validation with state-of-the-art tools:
Ramachandran plot analysis (PROCHECK)
- Stereochemical integrity
QMEAN scoring (SWISS-MODEL)
- Global model reliability
ProSA-web
- Z-score benchmarking against known structures
MolProbity
- All-atom geometry and clash detection
ERRAT
- Non-bonded interaction quality analysis
Our pipeline ensures that more than 90% of residues fall within favored regions of the Ramachandran plot, confirming structural reliability. Every project includes a detailed validation report, providing both numerical metrics and visual outputs to support confidence in the model.
How Protein Structures Are Predicted
Determining a protein’s structure can be done experimentally (X-ray crystallography, NMR, cryo-EM). However, these are expensive and time-intensive.
Therefore, computational protein structure prediction services offer a faster, lab-free alternative. For example, they include:
- Template-based modeling (homology modeling, threading)
- Ab initio methods for novel folds
- Deep learning approaches (AI/ML frameworks such as AlphaFold and RoseTTAFold)
- Hybrid pipelines combining refinement, quality scoring, and functional annotation
At Biointelix, we integrate these approaches into robust in-silico protein modeling services. Consequently, we provide accurate 3D protein models, functional annotations, and druggability insights directly from raw sequence data.
Beyond Prediction: Functional Insights
Our services extend beyond static protein structure prediction. We deliver functional context, including:
- Binding site annotation and pocket detection
- Conserved motif and active site identification
- Post-translational modification site prediction (glycosylation, phosphorylation, disulfide mapping)
- Molecular dynamics simulations (MD) to assess conformational stability under physiological conditions
- Protein–protein and protein–ligand interface analysis
These insights transform our models into actionable tools for structure-guided drug discovery, antibody design, and vaccine development.
Why Protein Structure Prediction Matters
Proteins are the molecular engines of life, and their structure ultimately determines their function.
Thus, predicting protein structure enables researchers to:
- Connect protein shape with molecular function.
- Identify druggable sites.
- Design vaccines and therapeutic antibodies.
- Model protein–ligand and protein–protein interactions.
The Four Levels of Protein Structure
Proteins are organized into four structural levels, each crucial for function:
- Primary Structure: The linear amino acid sequence. Even so, a single mutation can drastically alter folding and function.
- Secondary Structure: Local motifs such as α-helices and β-sheets, stabilized by hydrogen bonds.
- Tertiary Structure: The complete 3D folding of a polypeptide, shaped by hydrophobic, ionic, and disulfide interactions.
- Quaternary Structure: The assembly of multiple polypeptide chains into functional complexes (e.g., hemoglobin).
Because each level influences the next, understanding them is crucial for predicting interactions and identifying druggable sites. Furthermore, this knowledge creates the foundation for successful protein folding and structure prediction services.
Structure Determines Function
The shape of a protein dictates its activity. For example:
- Enzymes: Active sites fit substrates precisely; misfolding eliminates activity.
- Receptors: Ligand binding triggers cellular signaling.
- Antibodies: CDR loops form highly specific antigen-binding sites
- Structural Proteins: Collagen, keratin, and actin provide mechanical support.
Therefore, using computational protein modeling services allows researchers to connect protein structure with biological function in drug discovery and vaccine design. In addition, it supports innovation in diagnostics and therapeutics.
Protein Misfolding and Disease
Protein misfolding often leads to disease. For instance:
- Alzheimer’s: β-amyloid fibril accumulation
- Parkinson’s: α-synuclein aggregation
- Cystic fibrosis: ΔF508 mutation in CFTR
- Prion diseases: Infectious misfolded proteins
By comparing healthy and misfolded states, protein folding prediction services help researchers design better therapeutic strategies. Moreover, this highlights the critical role of accurate protein structure prediction in biomedical research.
Protein Structure in Drug Discovery
Structural knowledge significantly accelerates rational drug design. Specifically, it supports:
- Target identification: Locating druggable sites
- Structure-based design: Docking and screening candidate molecules
- Antibody engineering: Optimizing binding affinity
- Vaccine development: Mapping epitopes for immunogen design
For example, HIV protease inhibitors were developed using crystal structures — a landmark case in structure-guided drug discovery. Consequently, similar approaches are being used today with drug discovery protein modeling services that streamline pipelines and reduce costs.
Specialized Protein Structure Prediction Services
At Biointelix, we recognize that research often requires tailored structural solutions. Therefore, we offer specialized services, including:
Antibody Structure Prediction Services
- scFv, Fab, IgG, and single-domain antibodies with CDR loop refinement.
Membrane Protein Structure Prediction Services
- GPCRs, ion channels, and transporters, notoriously difficult to model experimentally
Protein–Protein Complex Structure Prediction Services
- Modeling interaction interfaces and docking multi-chain assemblies
Protein–Ligand Docking
- High-accuracy computational docking pipelines to model protein–ligand interactions. Our services help researchers identify binding affinities, optimize lead compounds, and accelerate drug discovery with reliable scoring functions and structural validation.
Binding Site Prediction
- Advanced binding site prediction services to identify potential druggable pockets on protein surfaces. We use AI-driven algorithms and structural bioinformatics tools to pinpoint ligand-binding regions, supporting rational drug design and precision medicine development.
Small Protein structure Prediction
- For novel folds and peptides lacking homologs.
Mutation and Stability Analysis Services
- Predicting how amino acid substitutions impact folding, binding, or developability
Each of these is supported by AI-based protein structure prediction services, structural bioinformatics services, and expert refinement. As a result, you gain confidence in results that are both predictive and actionable.
What You Receive
Each project delivers publication-ready results, including:
- Predicted 3D structures (PDB format)
- Refinement and validation reports (Ramachandran plots, QMEAN, ProSA, MolProbity)
- Binding site maps and druggability scoring (if requested)
- Publication-quality figures and 3D visualizations
- Optional molecular dynamics trajectories for advanced clients
This ensures you not only receive accurate models but also comprehensive documentation for research, patents, and publications.
Applications Across Life Sciences
Our Protein Structure Prediction Services support:
- Structure-based drug discovery
- Antibody and antigen engineering
- Enzyme redesign for industrial applications
- Structural genomics and functional annotation
- Vaccine candidate design and immune simulation
Consequently, Biointelix enables breakthroughs across multiple fields of life sciences. Moreover, our expertise ensures practical impact from computational modeling to experimental design.
Why Choose Biointelix for Protein Structure Prediction Services
- Deep Expertise: Specialists in protein modeling, docking, and vaccine design. Our published workflow on protein structure prediction, refinement, and validation demonstrates our commitment to scientific rigor (Palma, 2025; protocols.io).
- Custom Solutions: Tailored to your research needs
- Rapid Turnaround: Insights in days, not months
- Secure & Confidential: All data encrypted and protected
- Publication-Ready: High-quality figures and models
Therefore, by choosing Biointelix, you gain not only models but also a trusted partner in structural biology. Furthermore, our integrated approach ensures every project benefits from both speed and scientific rigor.
Get Started with Protein Structure Prediction Today
Unlock the full potential of computational protein structure prediction services. At Biointelix, we help design antibodies, explore therapeutic targets, and accelerate vaccine development. Moreover, our in-silico protein modeling services ensure that you move from raw sequence to actionable insights efficiently. As a result, you can make faster and smarter decisions in your research pipeline.
Contact us today for a free consultation to discuss your project and specific needs.
Recent Publications in Protein Structure Prediction and Modeling
To highlight the rapid progress in the field, here are some notable recent studies that demonstrate the growing applications of protein structure prediction and modeling:
(These external publications are provided as examples of the wider scientific advances in protein structure prediction and computational biology.)
References
Our published workflow in protein structure modeling and validation:
Marco Palma (2025). Workflow for Protein Structure Prediction, Refinement, and Validation. protocols.io
https://dx.doi.org/10.17504/protocols.io.dm6gpmz2dgzp/v1
Palma, M. (2025). Consensus-Guided Construction of H5N1-Specific and Universal Influenza a Multiepitope Vaccines. Biology, 14(10), 1327. https://doi.org/10.3390/biology14101327
